Computational Microscopy of SARS-CoV-2
Using Simulations to See the Unseen
Rommie Amaro
Professor, Department of Chemistry and Biochemistry
University of California - San Diego
About the Lecture
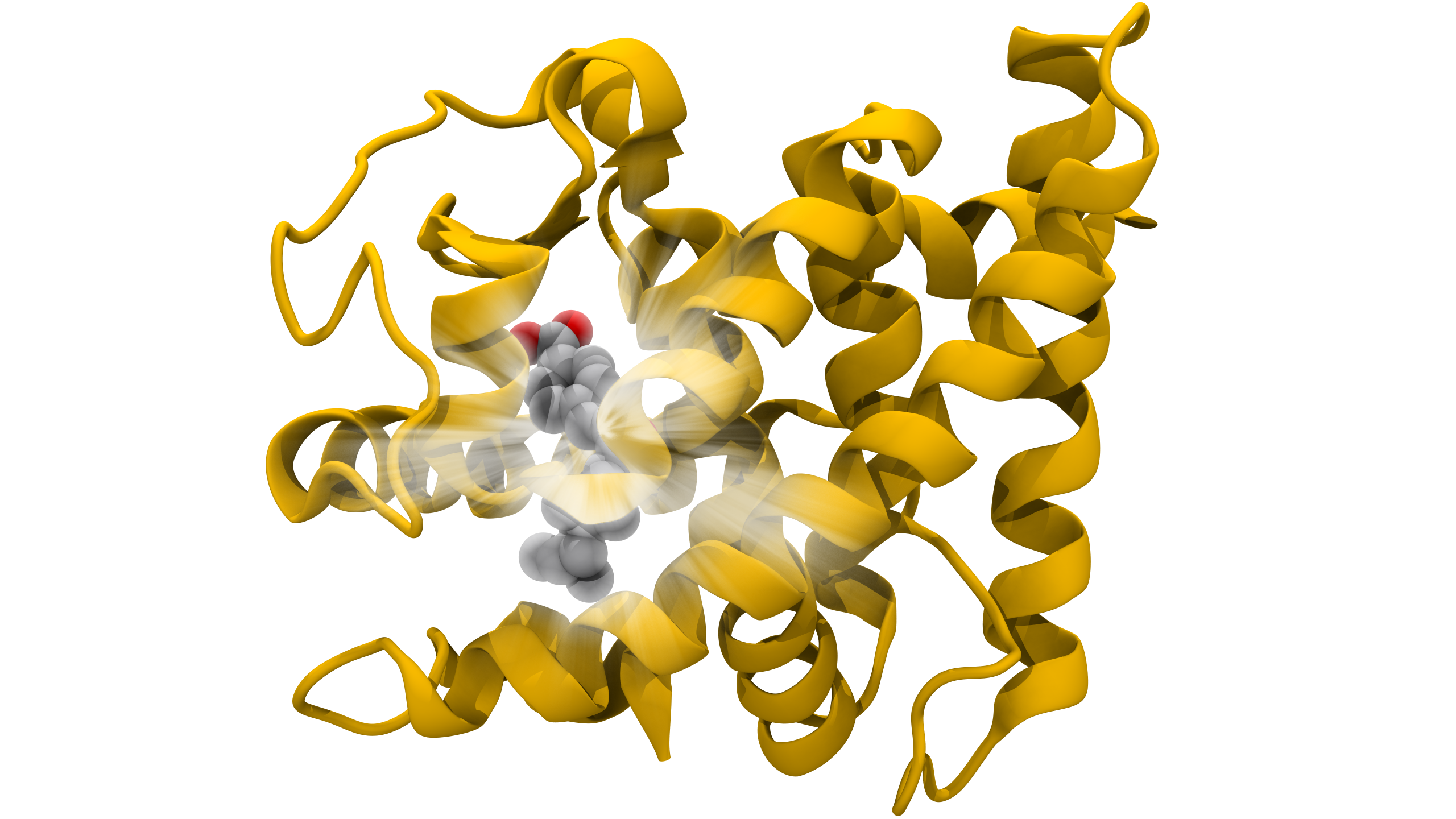
With advances in the acquisition of biological structures, researchers can resolve near-atomic resolution structures of viral components. These studies give valuable information about what these proteins look like, which is important for understanding infection processes and developing therapeutic vaccines and drugs. However, these datasets are subject to experimental limitations and, although they are immensely informative, they cannot provide a complete picture of these molecular machines and how they work. For example, experimental structure determinations often require altering a protein’s amino acid sequence to improve its expression, to trap it in a particular state or alter its biophysical properties. Regions that are highly dynamic are often removed because they limit resolution. Often biological components, such as lipid membranes and glycans, are difficult if not impossible to structurally resolve by available instrumentation and methods. Computational simulations are not subject to the same limitations as experiments, and often can be used to augment and extend experimental datasets. Biophysical molecular dynamics simulations are one such technique that allows researchers to resolve the structures of experimentally determined biomolecular machines back to their original state, and to add components that elude experimental characterization.
This lecture will discuss the use of computational microscopy to understand the SARS-CoV-2 virus in atomic detail, to better understand molecular recognition between the virus and its host cell receptors and antibody binding to the virus, and to advance the design of more effective antibodies, and the search for novel therapeutics.
The lecture will focus on studies of the SARS-CoV-2 spike protein, its glycan shield, its interactions with the human ACE2 receptor, and efforts to model the SARS-CoV-2 virion and the virions of the SARS-CoV-2 escape variants. The lecture will also discuss research that is dramatically revising current models of airborne transmission of respiratory viruses by providing never-before-seen atomic level views of the SARS-CoV-2 virus within a respiratory aerosol.
About the Speaker

Rommie Amaro is Professor and Endowed Chair in the Department of Chemistry and Biochemistry at the University of California, San Diego. Previously she was a member of the faculty of the Departments of Pharmaceutical Sciences, Computer Science, and Chemistry at UC – Irvine. Before joining the UC-Irvine faculty, Rommie did postdoctoral work at UC-San Diego, and she worked on condensed matter for Kraft Foods.
Rommie’s research is broadly concerned with the development and application of state-of-the-art computational methods to outstanding questions in molecular biophysics and drug discovery. In particular, she is working to develop multiscale simulation methods and novel modeling paradigms that scale from the level of atoms to the level of whole cells. She works closely with experimentalists to test these models and to use them in the design of empirical studies. And she is keenly interested in applying these methods to discover potential therapeutic agents.
Much of her research has been on influenza, Chlamydia, several less well studied diseases, and cancers. However, recently Rommie has been applying computational methods to understand the SARS-CoV-2 virus, how it binds and enters cells, and its mechanisms of action, with special attention to the spike protein.
Among other honors and awards, Rommie is the recipient of an NIH Kirschstein-National Research Service Award, an NIH New Innovator Award, a Presidential Early Career Award for Scientists, and an ACM Gordon Bell Special Prize for work on SARS-CoV-2. She has also been appointed to an endowed Professorship in the Department of Chemistry and Biochemistry at UC – San Diego.
Rommie earned a BS in Chemical Engineering and a PhD in Chemistry at the University of Illinois – Urbana-Champaign.
Minutes
On November 5, 2021, by Zoom webinar broadcast on the PSW Science YouTube channel, President Larry Millstein called the 2,448th meeting of the Society to order at 8:02 p.m. EDT. He welcomed new members, and the Acting Recording Secretary read the minutes of the previous meeting.
President Millstein then introduced the speaker for the evening, Rommie Amaro, Professor and Endowed Chair in the Department of Chemistry and Biochemistry at the University of California, San Diego. Her lecture was titled, “Computational Microscopy of SARS-CoV-2: Using Simulations to See the Unseen.”
Amaro’s research group, among others, has developed a technique called molecular dynamic simulations, which she characterizes as a computational microscope. The technique uses computing to synthesize diverse data sources to produce detailed three-dimensional models of biological systems that are then “brought to life” by using basic laws of physics. Her group has used this technique to use different structural data sets with glycomics, to approximate the SARS-CoV-2 system and reduce it to basic mechanical elements. By then applying Newton’s equation of motion, Amaro’s group builds a dynamical understanding of the system’s movement over time.
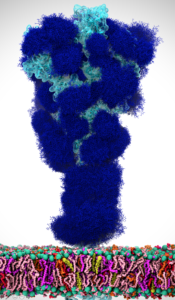
In February 2020, the product of seven years research, Amaro’s team presented a paper on a molecular dynamics simulation of the outer envelope of the influenza virus. Contemporaneous with that publication, a research collaboration with the U.S. National Institutes of Health published the first cryo-electron microscopy structure of the SARS-CoV-2 (also known as 2019-nCoV) spike protein. Amaro then described that structure and its modeling in detail.
Spike proteins sit on the virus surface and thus are the first contact the virus has with human cells. By using their computational microscope to fill in gaps in the NIH team’s model, Amaro’s team was able to “see” the protein’s complete spike structure and its membrane composition.
The speaker showed a “slice” of the membrane that her team modeled, illustrating the cytoplasmic tail that sits inside the membrane. She then showed and described animations of the virus structure subjected to Newtonian physics. The simulation showed significant “wiggling and jiggling,” explaining why crisp imagery of the virus proved so elusive.
Amaro said that while the images were computationally expensive, they allowed researchers to compute properties that link directly to lab-testable experiments. From these experiments, scientists were able to confirm what the full-length, fully glycosylated spike protein looks like – including sugar linkages that scientists could not previously see. Amaro then detailed their specific findings. This information is crucial for SARS-CoV-2 vaccine and drug design.
In the absence of conferences, Amaro said that social media quickly became the means through which she communicated with other scientists about their research. By Twitter, she was able to meet Elisa Fadda of Maynooth University in Ireland, with whom she collaborated to develop an experimentally testable hypothesis that SARS-CoV-2 glycans had purposes beyond shielding.
Also by Twitter, Jason McClellan (co-inventor of the Moderna vaccine) responded to one of Amaro’s spike protein videos. As a result, McClellan’s lab developed a bio-interferometry assay that showed spike protein mutations supporting Amaro’s hypothesis that the virus’s glycans participated in the mechanism of infection.
The speaker then described her team’s work on the fusion of the virus’s two membranes. They partnered with Lillian Chong at the University of Pittsburgh, who developed a method called weighted ensemble molecular dynamics. The method is a statistically rigorous way to overcome the computing barriers to standard molecular dynamics and simulate the virus’s spontaneous membrane opening. Whereas standard molecular dynamics would take more than 800 years to determine one pathway, Chong’s method accomplished 310 pathways in 23 days. These weighted ensemble trajectory simulations are in agreement with the ManifoldEM results of PSW member Joachim Frank’s laboratory, conducted by graduate student Evan Seitz. The resulting conclusion is that a glycan gate controls opening of the SARS-CoV-2 spike protein.
Amaro also studied the ACE-2 receptor. Her team found the receptor was “surprisingly flexible,” facilitating the mechanical process of membrane fusion.
The dynamics developed from the weighted ensemble simulations have now been fed into a deep learning method to make simulations more efficient. Experimentalists have since uncovered SARS-CoV-2 morphology, including the size of the membrane envelope, and both the number and distribution of spike proteins on the surface of the virus. From those findings, Amaro’s team has been able to simulate methods for building the virus’s membrane envelope.
Amaro’s laboratory work has now been recognized by the Gordon Bell Special Prize for High Performance Computing-Based COVID-19 Research. Ongoing work continues through worldwide collaboration and information sharing, using freely available, open access community software.
The speaker then answered questions from the online viewing audience. After the question and answer period, President Millstein thanked the speaker, made the usual housekeeping announcements, and invited guests to join the Society. President Millstein adjourned the meeting at 10:23 p.m.
Temperature in Washington, D.C.: 10° C
Weather: Clear
Concurrent Viewers of the Zoom and YouTube live stream, 46 and views on the PSW Science YouTube and Vimeo channels: 189.
Respectfully submitted,
James Heelan, Recording Secretary