Living Measurement Systems and Minimal Cells
Engineering Cellular Life for Understanding and Applications
Elizabeth Strychalski
Group Leader, Cellular Engineering Group
National Institute of Standards & Technology
About the Lecture
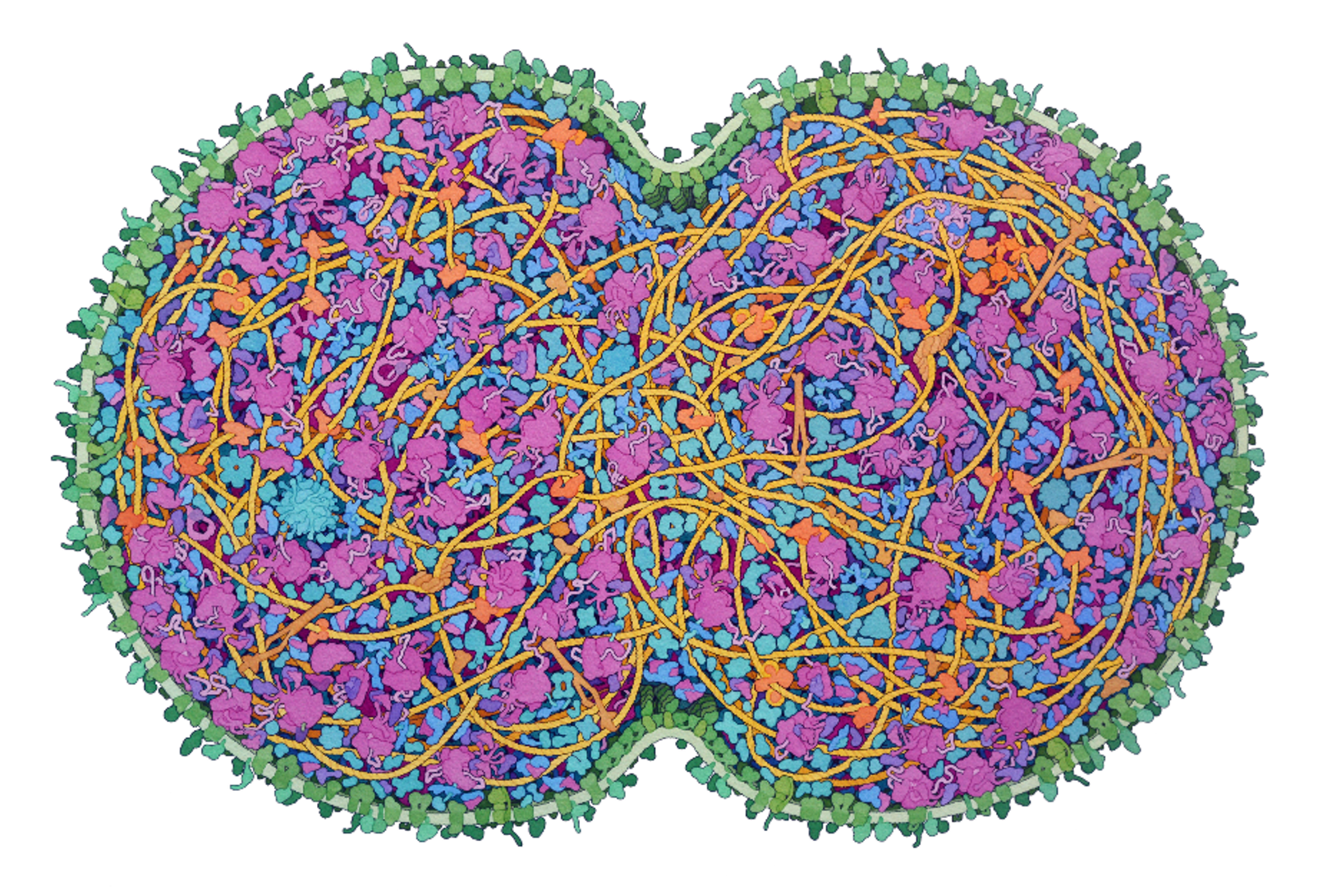
Measurement tools made from biological parts—living measurement systems—offer a compelling opportunity to better control and engineer biological systems in a way that takes full advantage of naturally-evolved biology and human capabilities for engineering living systems.
This lecture will begin by summarizing efforts at the National Institute of Standards and Technology to build living measurement systems. The efforts include directed evolution for engineering protein sensors, cellular computation using continuously transcribed RNA strand displacement circuits, and measurement assurance for engineering biology chassis.
The lecture will then turn to a deeper consideration of genomically minimal cells, whose creation represents a tour de force of modern biology. A review of the underlying capabilities leading to the minimal cell offers suggestions for further optimization and future possibilities for other minimized cells. Despite its well-understood genome, the minimal cell retains genes of unknown function. Absent a comprehensive understanding of the role of every gene in the minimal cell, opportunities remain to bridge genotype and phenotype using physical models for insight into gene function and to inform engineering of cellular function.
Further Information
(1) Fundamental behaviors emerge from simulations of a living minimal cell, Thornburg et al. (2022): Cell 185 (2) pgs 345-360. https://www.sciencedirect.com/science/article/pii/S0092867421014884?via%3Dihub
(2) Genetic requirements for cell division in a genomically minimal cell, Pelletier et al. (2021): Cell 184 (9) pgs 2430-2440. https://www.sciencedirect.com/science/article/pii/S0092867421002932?via%3Dihub
(3) Design and synthesis of a minimal bacterial genome, Hutchison et al. (2016): Science 351 (6280). doi 10.1126/science.aad6253. https://www.science.org/doi/10.1126/science.aad6253?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%20%200pubmed
Additional information is available on the group website at: https://www.nist.gov/mml/bbd/cellular-engineering-group
About the Speaker

Elizabeth Strychalski is Group Leader of the Cellular Engineering Group at the National Institute of Standards and Technology (“NIST”). Before joining NIST she was Program Manager in the Biological Technologies Office at DARPA.
Elizabeth founded the Cellular Engineering Group and works to provide a foundation of measurements to support the design and control of engineered function in living systems. In her previous work at DARPA, Elizabeth managed a portfolio to aggressively advance experimental and theoretical research at the interface of synthetic biology, control engineering, nanobiotechnology, and fluidic devices.
Among other honors and awards, Elizabeth is the recipient of the University of Rochester’s Harry W. Fullbright Prize and its Janet Howell Clark Award, and she received the Maryland Academy of Sciences Young Scientist Award.
Elizabeth earned a BS in Physics and Astronomy and a BA in Religious Studies at the University of Rochester, and an MA and PhD in Physics at Cornell University
Minutes
On April 22, 2022, from the Powell Auditorium of the Cosmos Club in Washington, D.C, and by Zoom webinar broadcast on the PSW Science YouTube channel, President Larry Millstein called the 2,457th meeting of the Society to order at 8:02 p.m. ET. He welcomed new members, and the recording secretary read the minutes of the previous meeting.
President Millstein then introduced the speaker for the evening, Elizabeth Strychalski, Group Leader of the Cellular Engineering Group at the National Institute of Standards & Technology (NIST). Her lecture was titled, “Living Measurement Systems and Minimal Cells: Engineering Cellular Life for Understanding and Applications.”
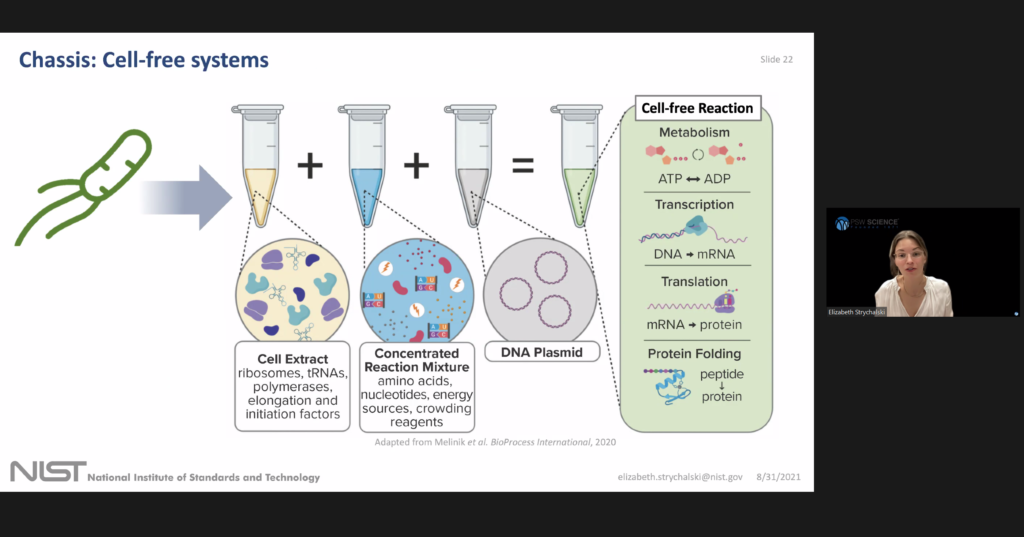
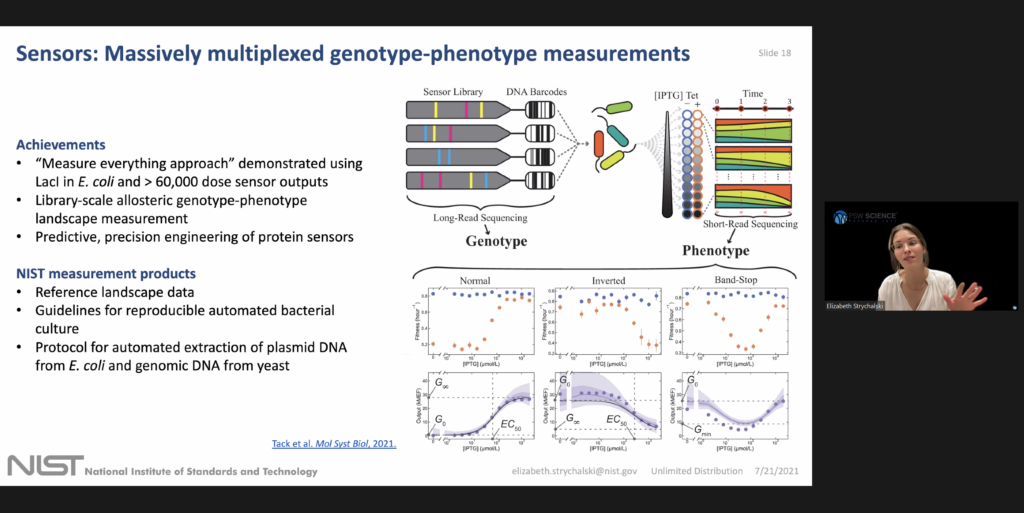
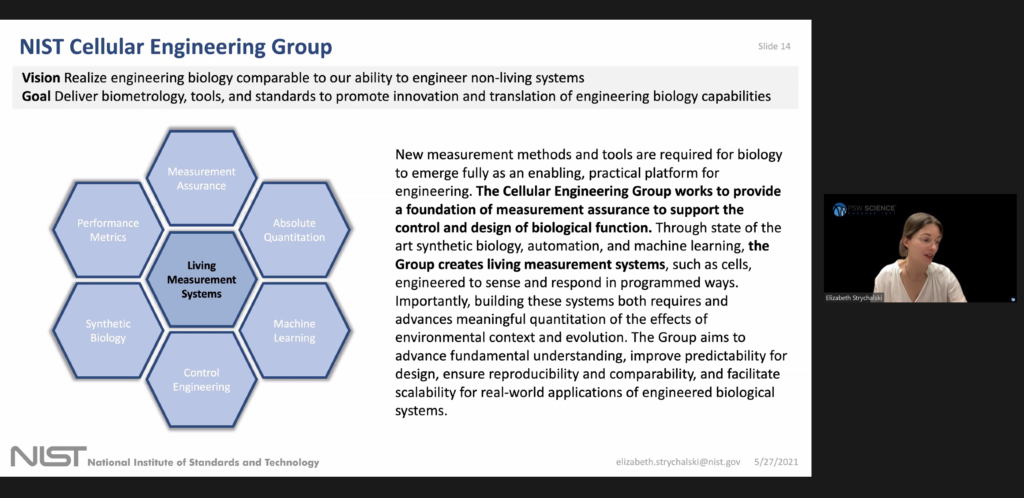
Strychalski began by introducing three areas that her cellular engineering group is working on: advance living measurement systems, or sensors, computation, and chassis. She explained that the living measurement systems foundry is part of NISTs larger push to modernize and automate the biological labs at NIST.
Strychalski highlighted that her lab is actively sharing their experience and advice on automating synthetic biology protocols by publishing a technical manuscript detailing protocols, sharing information methods on GitHub, and they are a member of the global biofoundry alliance.
The speaker then moved on to the topic of minimal cellular life beginning with her goal, which is to be able to engineer a platform using minimal cells for developing and testing living measurement systems.
The development of the minimal cell at the heart of the lecture was led by John Glass, at the J. Craig Venture Institute, or JCVI. JCVI develops minimal cells with a top-down approach by removing parts from an already functioning cell until it just barely meets the definition of life. Three key capabilities are leveraged to minimize cellular genomes: genome synthesis, genome transplantation, and a Design-Build-Test workflow. These capabilities were applied to the organism, Mycoplasma mycoides.
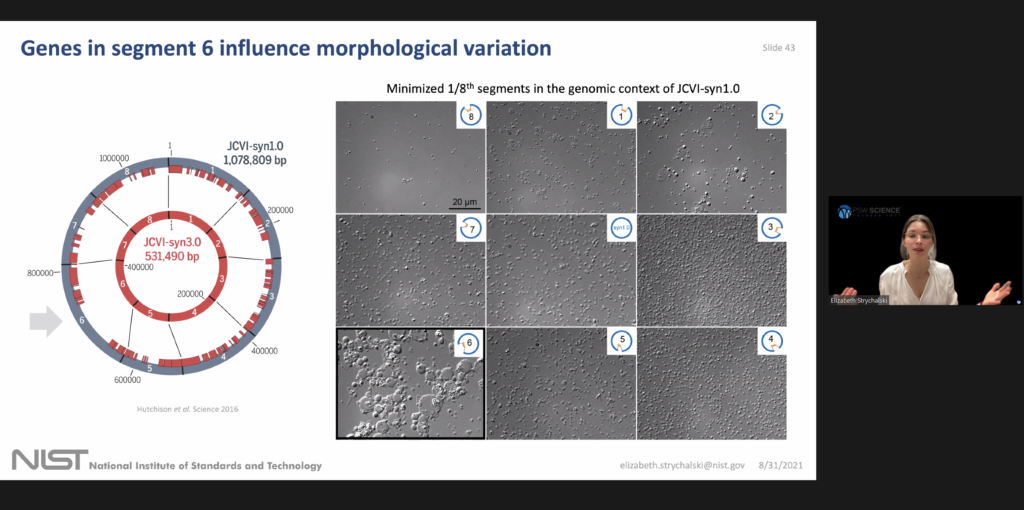
Strychalski explained the origin of JCVI-syn1.0 as being the Mycoplasma mycoides genome transplanted into a related species, capricollum. Strychalski explained that the minimal cell was developed using eight genome segments. Eight was chosen, she admits, mostly arbitrarily.
She then compared the growth of JCVI-syn 1.0 and JCVI-syn 3.0. JCVI-syn 3.0 is a subset of the genome 1.0, and represents the first realization of the concept of minimal cells as it was originally proposed.
Practical problems arise with trying to study and engineer cells that don't have reproducible characteristics of the single cell scale. To address the difficulties of measuring with an electron micrograph, which requires fixing cells which kills them, they developed microfluidic chemostats for measuring the dynamics of single cell growth and investigating what genes are required for normal morphology.
They restricted the search from the 428 genes that were removed from syn 1.0 to make syn 3.0 have only 76 genes missing from the minimized segment six. By focusing on a segment 6 they developed a strain called JCVI-syn3A. It has 493 genes and 19 genes not in the minimal version of segment 6 and it shows nearly normal cell division. She emphasized syn 3a because it's the most widely used strain among the dozens of labs now doing research with this minimal cell.
The team refined the search to finding which of the 19 genes was most essential to controlling cell division. They hypothesized that cell division depended on a gene cluster that is part of a very highly conserved division and cell wall cluster. They found seven genes known for cell division and four genes that encode what they think are membrane-associated proteins.
The speaker then considered, as a physical object, the question of what mechanical forces are acting on the membrane that could drive constriction incision during cell division. The lab is looking into which parameters these genes might depend on in this genomically minimal context. This will hopefully give information on what these gene products are doing and help realize the vision for understanding what every gene in the cell is doing.
The speaker then reviewed recent publications and preprints involving JCVI-syn3A and spoke about what is next with minimal cells. She advocated for an international community called Build a Cell.
Next the question and answer period began. One of the members asked how the speakers work was different than that of Eric Winfrey's, or if it builds upon his work. The speaker responded that his work was part of the inspiration for their RNA circuits and noted that the difference is that, in general, for DNA computations you typically make the different molecular components of the computation in separate tubes and then have to mix them together they react once and then that's it you're done whereas the speaker’s lab recruits the machinery of life to make the parts all together in situ.
A question from a member participating through YouTube asked if there was any work being done to improve the ethanol production from switchgrass metabolizing. The speaker commented more generally on the cell-based biomanufacturing, which is a NIST supported area. She responded that there may be some opportunity for using minimal cells that have been engineered to function as living measurement systems and perhaps co-culture them or use them in some other way as part of a monitoring system in these biomanufacturing processes so they can be competitive against petroleum processes.
After the question and answer period, President Millstein thanked the speaker, made the usual housekeeping announcements, and invited guests to join the Society. President Millstein adjourned the meeting at 9:57 p.m.
Temperature in Washington, D.C.: 20° C
Weather: Partly Cloudy
Number of streaming online viewers: 37, and views on the PSW Science YouTube and Vimeo channels in the first two weeks of the lecture: 155.
Respectfully submitted,
Cameo Lance, Director of Special Projects
Highlights